Predictor
ℹ️ Variable-Length Sequence Processing
EnDeep4mC can process DNA sequences of any length (20-100,000 bp) using the following pipeline:
1. Length Standardization
All sequences are standardized to 41bp windows through appropriate padding or segmentation
2. Cytosine Filtering
Only windows with cytosine (C) at the center position are analyzed for 4mC prediction
3. Sliding Window Analysis
Long sequences are analyzed using overlapping 41bp windows with 1bp step size
4. Ensemble Prediction
Each 41bp window is analyzed by CNN, BLSTM, and Transformer ensemble
Note: For sequences longer than 41bp, predictions are averaged per cytosine position to provide robust methylation likelihood scores.
Model Architecture Details
Table 1. Architecture and Hyperparameters of Base Deep Learning Models
Table 2. Configuration of Ensemble Learning Framework
Model Architecture Diagram
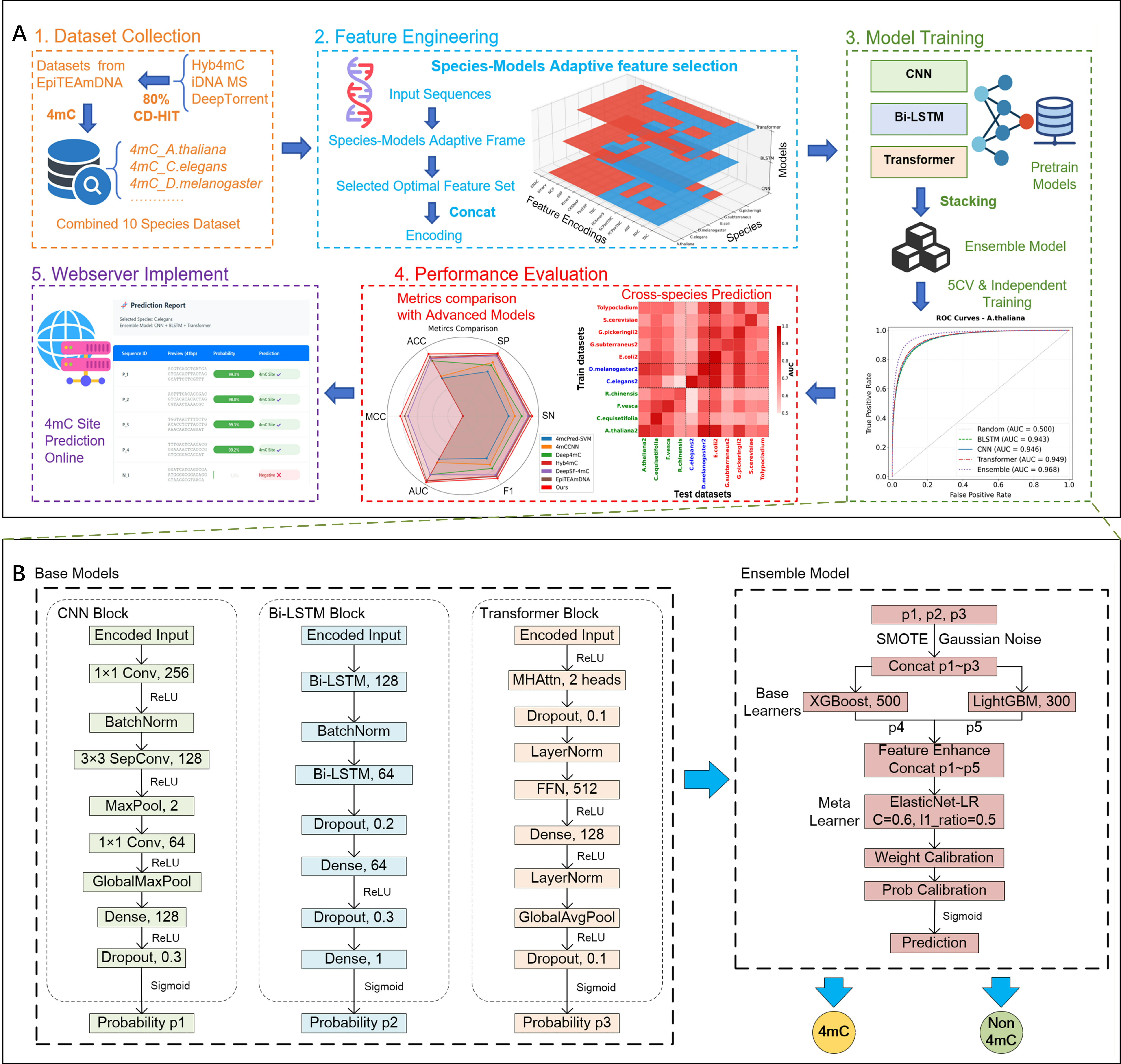
Integrated Deep Learning Architecture with Dual-Adaptive Encoding System
Example FASTA Format
>Sample_Sequence_1 (41bp) ATGCTAGCTAGCTAGCTAGCTAGCTAGCTAGCTAGCTAGCTAGC >Sample_Sequence_2 (50bp) CGATCGATCGATCGATCGATCGATCGATCGATCGATCGATCGATCGATCGA >Sample_Sequence_3 (30bp) ATCGATCGATCGATCGATCGATCGATCGAT
Variable-Length Examples: The webserver accepts sequences of any length (20-100,000 bp). Short sequences will be standardized to 41bp, and long sequences will be analyzed using sliding windows.